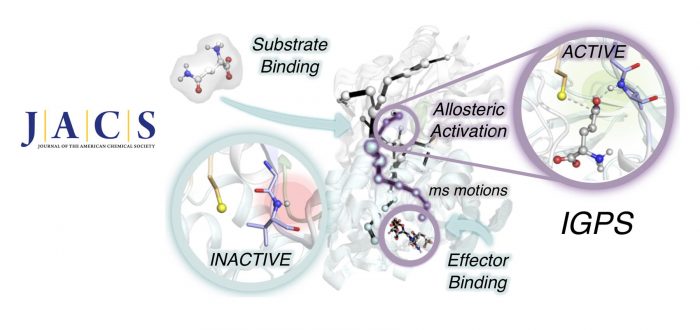
Proteins reshape their function in response to environmental changes through allosteric regulation. Allostery is the process in which two distinct sites within a protein or protein complex are functionally coupled. In allosterically regulated enzymes, the binding of a molecule (allosteric effector) at a distal site alters the thermodynamic and/or kinetic parameters of the catalytic reaction at the active site. The transfer of chemical information between the two energetically coupled sites is mediated by structural and/or dynamical changes that generally make accessible the preorganized active site conformation characteristic of the enzyme active state. To attain such a catalytically competent state, effector binding finely tunes the enzyme dynamic conformational ensemble by reshaping the relative populations of the conformational states and/or the time scales of structural fluctuations and conformational transitions.
Complete bidirectional communication between distal sites occurs at the ternary complex, i.e., when both the effector and substrate are bound at their respective sites, and propagates through dynamic networks of inter- and intramolecular interactions. Capturing the time evolution of the allosteric activation of enzymes toward the formation of the ternary complex involves deciphering the interplay of fast and slow conformational dynamics coupled to effector and substrate binding.The transient nature of the ternary complex and the usually slow millisecond time scale allosteric transition in enzymes undergoing turnover hampers the structural and dynamic characterization of allosteric mechanisms and the identification of functionally relevant states. It is therefore not surprising that the allosterically active state remains hidden for several enzymes.
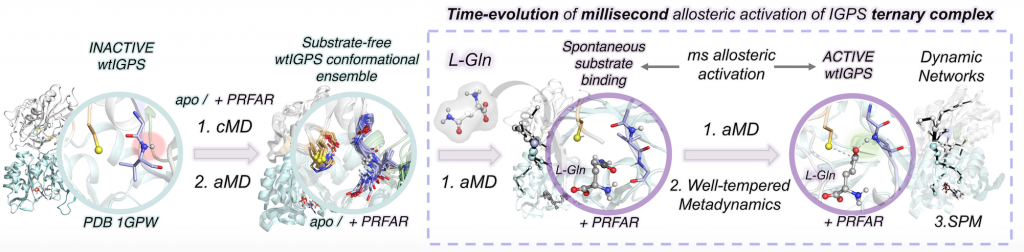
In this work, we combine extensive molecular dynamics simulations, enhanced sampling techniques (accelerated molecular dynamics and metadynamics), and dynamical networks (shortest path map) to describe the allosteric activation of imidazole glycerol phosphate synthase (IGPS) from the substrate-free form to the active ternary complex. IGPS is a heterodimeric bienzyme complex whose HisH subunit is responsible for hydrolyzing glutamine and delivering ammonia for the cyclase activity in HisF. Despite significant advances in understanding the underlying allosteric mechanism, essential molecular details of the long-range millisecond allosteric activation of IGPS remained hidden.
Without using a priori information of the active state, our simulations uncover how IGPS, with the allosteric effector bound in HisF, spontaneously captures glutamine in a catalytically inactive HisH conformation, subsequently attains a closed HisF:HisH interface, and finally forms the oxyanion hole in HisH for efficient glutamine hydrolysis. We show that the combined effector and substrate binding dramatically decreases the conformational barrier associated with oxyanion hole formation, in line with the experimentally observed 4500-fold activity increase in glutamine hydrolysis. The allosteric activation is controlled by correlated time-evolving dynamic networks connecting the effector and substrate binding sites. This computational strategy tailored to describe millisecond events can be used to rationalize the effect of mutations on the allosteric regulation and guide IGPS engineering efforts.
This work has been carried out by IQCC researchers Carla Calvó-Tusell, Miguel A. Maria-Solano (now at Ewha Womans University, Republic of Korea), Sílvia Osuna, and Ferran Feixas, and was recently published in Journal of the American Chemical Society:
C. Calvó-Tusell, M.A. Maria-Solano*, S. Osuna*, and F. Feixas
“Time Evolution of the Millisecond Allosteric Activation of Imidazole Glycerol Phosphate Synthase”
J. Am. Chem. Soc. 2022, 144, 7146-7159
DOI: 10.1021/jacs.1c12629
Girona, April 29, 2022
For more info: gestor.iqcc@gmail.com