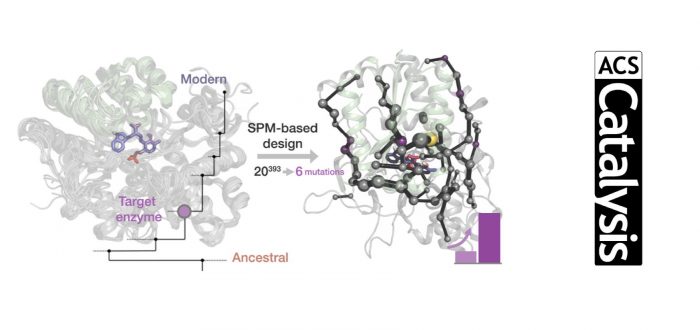
Allostery is a central mechanism for the regulation of multi-enzyme complexes. The mechanistic basis that drives allosteric regulation is poorly understood but harbors key information for enzyme engineering. In the present study, we focus on the tryptophan synthase complex that is composed of TrpA and TrpB subunits, which allosterically activate each other. Specifically, we develop a rational approach for identifying key amino acid residues of TrpB distal from the active site. Those residues are predicted to be crucial for shifting the inefficient conformational ensemble of the isolated TrpB to a productive ensemble through intra-subunit allosteric effects.
The experimental validation of the conformationally-driven TrpB design demonstrates its superior stand-alone activity in the absence of TrpA, comparable to those enhancements obtained after multiple rounds of experimental laboratory evolution. This work evidences that the current challenge of distal active site prediction for enhanced function in computational enzyme design has become within reach.
This work is the result of a nice collaboration between the University of Regensburg: Thomas Kinateder, Prof. Reinhard Sterner (Sterner lab) and our institute: Dr. Miguel Ángel Maria-Solano, Prof. Sílvia Osuna, and was published recently in ACS Catalysis:
M.A. Maria-Solano, T. Kinateder, J. Iglesias-Fernández, R. Sterner, and S. Osuna
“In Silico Identification and Experimental Validation of Distal Activity-Enhancing Mutations in Tryptophan Synthase”
ACS Catal. 2021, 11, 13733-13743
DOI: 10.1021/acscatal.1c03950
Girona, November 19, 2021
For more info: gestor.iqcc@gmail.com