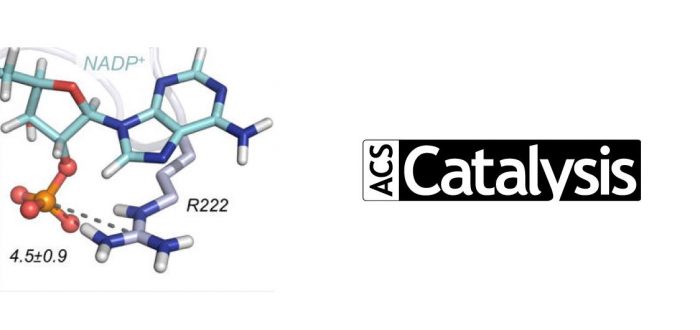
The development and application of a novel in vivo selection method enabled the testing of more than >106 variants of formate dehydrogenase (FDH) to switch its cofactor specificity in one single round of protein directed evolution, using multiple mutational sites selected based on previous studies and initial computational modeling. Due to this new approach, key cooperative epistatic effects between several amino acid positions arose, which were characterized experimentally by enzyme kinetics and rationalized computationally through advanced molecular dynamics simulations.
The work involved the collaboration between four different labs: Liliana Calzadiaz-Ramirez,# Steffen Lindner and Arren Bar-Even from the Max Planck Institute of Molecular Plant Physiology; Gabriele Stoffel# and Tobias Erb from the Max Planck Institute of Terrestrial Microbiology; Carlos Acevedo-Rocha from Biosyntia; and Carla Calvó-Tusell,# Sílvia Osuna and Marc Garcia-Borràs from the IQCC, UdG.
#these authors contributed equally as co-first authors
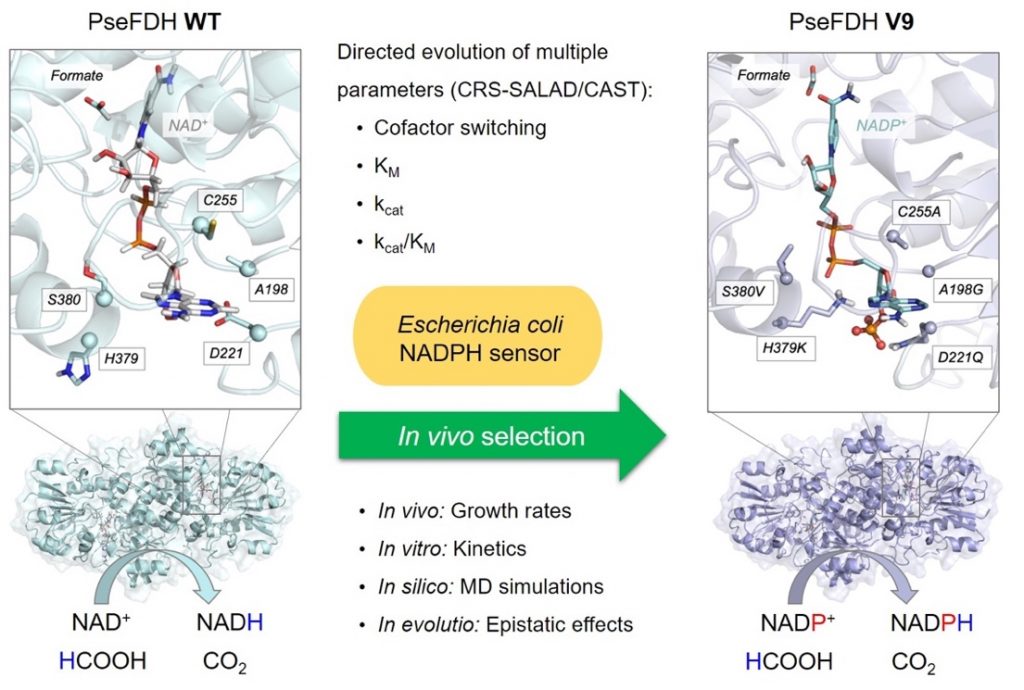
This new multidisciplinary approach led to engineered FDH variants displaying the highest activities and specificities reported thus far. While such new enzymes could be employed in various applications to use formate for cofactor recycling or as feedstock for the biosynthesis of high added-value products, the strategy proposed in the present work can reduce the time and cost for engineering cofactor specificity in other oxidoreductases, thus expanding the biocatalytic toolbox in protein and metabolic engineering within the synthetic biology community.
About the study:
The efficient regeneration of cofactors is vital for the establishment of biocatalytic processes. Formate is an ideal electron donor for cofactor regeneration due to its general availability, low reduction potential, and benign byproduct (CO2). However, FDHs are usually specific to NAD+, such that NADPH regeneration with formate is challenging. Previous studies reported naturally occurring FDHs or engineered FDHs that accept NADP+, but these enzymes show low kinetic efficiencies and specificities. Here, the power of natural selection has been harnessed to engineer FDH variants to simultaneously optimize three properties: kinetic efficiency with NADP+, specificity toward NADP+, and affinity toward formate. By simultaneously mutating multiple residues of FDH from Pseudomonas sp. 101, which exhibits practically no activity toward NADP+, a library of >106 variants was generated. This library was then introduced into an E. coli strain that cannot produce NADPH. By selecting for growth with formate as the sole NADPH source, several enzyme variants that support efficient NADPH regeneration were isolated. It was found that the kinetically superior enzyme variant, harboring five mutations, has 5-fold higher efficiency and 14-fold higher specificity in comparison to the best enzyme previously engineered, while retaining high affinity toward formate. Extensive computational modelling revealed the contribution of each mutation to the superior kinetics of this variant. Additionally, nonadditive epistatic effects were determined to improve multiple parameters simultaneously. The present work demonstrates the capacity of in vivo selection, based on rationally designed libraries, to identify highly proficient enzyme variants carrying multiple mutations which would be almost impossible to find using conventional screening methods.
The computational part of the project has been carried out by PhD student and co-first author Carla Calvó-Tusell. It has specifically focused on understanding at the atomic level why the wild-type variant (WT) is not active towards NADP+ but is highly efficient towards its natural cofactor NAD+, and how the specific mutations selected through this massive in vivo screening method reversed the situation. Initial MD simulations on the WT enzyme were used to select mutational hotspots for designing libraries for the laboratory directed evolution, together with previously published data.
After in vivo laboratory evolution, the best variants characterized in the laboratory were computationally modeled. Using MD simulations, and considering both natural and unnatural cofactors bound in evolved variants, it was possible to determine the individual and cooperative role of the different mutations introduced in the FDH active site to selectively and efficiently accept NADP+ instead of NAD+. These analyses showed that a remodeling of the binding pocket shape has taken place, and that also the polarity of the active site has been modified to stabilize the unnatural cofactor in a catalytic conformation.
Using theozyme (“theoretical enzyme”) truncated model calculations, the optimal geometry of the transition step (TS) for the hydride transfer step from the formate to the cofactor was determined, and this geometry was used as a reference to analyze the catalytic efficiency of the enzyme using MD data. These atomistic descriptions allowed to get a full and complete picture on the specific role of each new introduced mutation, rationalizing the experimental kinetic data of the different variants explored. Computational modelling provided very useful information about the enzyme catalytic machinery function, enzyme substrate / cofactor recognition mechanisms, and how cooperativity between newly introduced mutations emerged during in vivo evolution.
The study was recently reported in ACS Catalysis:
L. Calzadiaz-Ramirez, C. Calvó-Tusell, G.M.M. Stoffel, S.N. Lindner, S. Osuna, T.J. Erb, M. Garcia-Borràs, A. Bar-Even, and C.G. Acevedo-Rocha
“In Vivo Selection for Formate Dehydrogenases with High Efficiency and Specificity toward NADP+ ”
ACS Catal. 2020, [], 7512-7525
DOI: 10.1021/acscatal.0c01487
Girona, July 5, 2020
For more info: sec.iqcc@udg.edu