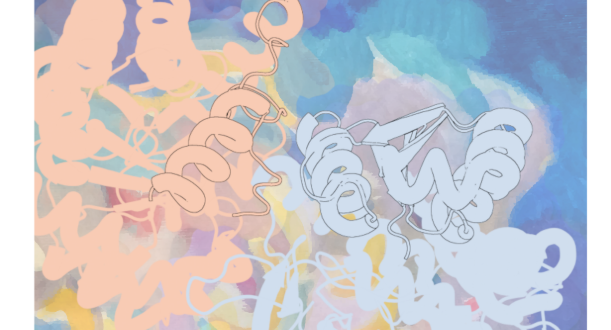The Protein Sci. journal features on its front cover of the December 2025 issue the research article “A naturally occurring standalone TrpB enzyme provides insights into allosteric communication within tryptophan synthase” by Prof. Sílvia Osuna in collaboration with Prof. Reinhard Sterner (University of Regensburg).

The study of enzymes continues to reveal new layers of complexity in how these sophisticated molecular catalysts achieve their remarkable efficiency, specificity and evolvability. Their catalytic prowess emerges from an intricate interplay of structural organization, conformational dynamics, evolutionary optimization, and allosteric regulation. This special issue of Protein Science showcases cutting-edge studies that advance our understanding across multiple frontiers of enzyme science, from fundamental mechanisms of adaptation and regulation to practical applications in biotechnology and sustainability.
An important frontier in enzyme research involves understanding how proteins adapt to extreme environments while maintaining catalytic function. The structural basis of cold adaptation represents a particularly fascinating example of molecular evolution under selective pressure. Work by Yang et al on bidomain cellulases, demonstrate how domain architectures and linkers can be fine-tuned to enhance activity at low temperatures. Their discovery that domain separation correlates with catalytic turnover at 10°C through the “linker spacer effect” illustrates how subtle changes in protein dynamics can yield substantial functional improvements, offering valuable insights for engineering energy-efficient industrial biocatalysts.
The challenge of enzyme optimization extends beyond natural adaptation to include rational design strategies that overcome inherent catalytic limitations. Product inhibition, a common bottleneck in enzyme biotechnology, represents a prime target for innovative engineering approaches. The work by Zheng et al. on DszB desulfinase demonstrates how synthetic modifications—in this case, strategic glycosylation—can alleviate conformational constraints that limit catalytic turnover. Their approach to mitigating product inhibition through enhanced loop flexibility exemplifies how understanding the molecular basis of catalytic limitations can guide targeted interventions with direct applications in biodesulfurization technologies.
The sophistication of enzyme activity modulation is evident in allosteric systems, where distant sites communicate to coordinate catalytic activity. The investigation by Kinateder and colleagues of stand-alone tryptophan synthase variants illuminates the molecular basis of allosteric communication, revealing how specific residue networks can modulate the conformational landscapes that govern catalytic competence. Their work demonstrates that allosteric effects can be largely context-independent, providing design principles for engineering cooperative enzyme systems.
The multi-domain architectures central to many of these catalytic innovations reflect fundamental evolutionary processes that have shaped enzyme complexity. At the intersection of structure and evolution, the relationship between protein architecture and functional diversification continues to yield fundamental insights. The perspective by Muthahari et al. expands our understanding of how complex proteins emerge through duplication and fusion events, highlighting fusion as a critical yet underappreciated driver of protein evolution. This evolutionary lens provides essential context for understanding the modular organization observed in many contemporary enzymes and offers principles for designing proteins with enhanced evolvability.
Collectively, these (and other) studies included in this issue illustrate the current state of enzyme science, where advanced computational methods, structural biology, and evolutionary perspectives converge to address both fundamental questions about catalytic mechanisms and practical challenges in biotechnology. These insights promise to accelerate both the development of novel pharmaceuticals against challenging therapeutic targets, as well as next-generation biocatalysts for applications ranging from sustainable manufacturing to environmental remediation.
The corresponding paper was published recently:
Thomas Kinateder, Lukas Drexler, Cristina Duran, Sílvia Osuna*, Reinhard Sterner*
A naturally occurring standaloneTrpB enzyme provides insights into allosteric communication within tryptophan synthase
Protein Science, 2025, 34, e70103
DOI: 10.1002/pro.70103
Girona, February 9th, 2026
For more info: ges.iqcc@udg.edu