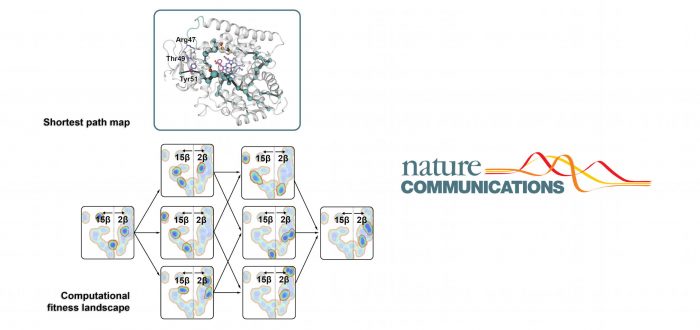
The systematic construction of the so-called protein fitness landscapes and the quantification of non-additive effects observed by mutations (known as epistasis) are rarely reported. However, they provide fundamental insights into the mechanisms of laboratory and natural evolution. When reported, unfortunately, these studies usually consider only a single enzyme trait of a limited number of protein families, resulting in an incomplete picture of the evolution of multiple protein functions with little or no reference to possible effects caused by cooperative (positive) or deleterious (negative) epistasis.
In this collaboration project between groups located in Germany, Denmark, the UK, China, Australia, and IQCC the first multiparametric fitness landscape of a cytochrome P450 is reported and analyzed in depth. Cytochromes P450 are a superfamily of enzymes containing heme as a cofactor that function as monooxygenases. Using a P450-BM3 variant as the regio- and stereoselective catalyst in the pharmaceutically valuable CH-activating hydroxylation of a steroid (testosterone), all possible trajectories connecting the parental enzyme with the selective mutant were individually constructed and thoroughly analyzed by experimental and computational approaches.
Cooperative and deleterious epistatic influences were quantified with a newly developed computational program, revealing pervasive positive epistatic effects on all parameters along the six possible pathways. Lorenzo D’Amore, Dr. Marc Garcia-Borràs, and Prof. Sílvia Osuna explored by advanced Molecular Dynamics (MD) simulations together with QM calculations that these synergistic effects are modulated by long-range interactions in loops, helices and beta-strands which cause dramatic conformational changes affecting activity and selectivity traits of the enzyme. An analysis of the conformational dynamics unveils the mechanism of epistasis in which there are direct effects between mutations, but no direct interaction between the substrate and the mutations.
The unique information derived from this work can be expected to aid future protein engineering focused on P450-catalyzed oxidative hydroxylation of steroids and of other pharmaceutically significant substrates. This work can also inspire other researchers in the biocatalysis community to engage in the study of epistasis in other enzyme families particularly when multiple parameters are investigated.
The paper was published recently in Nature Communications:
C.G. Acevedo-Rocha, A. Li, L. D’Amore, S. Hoebenreich, J. Sanchis, P. Lubrano, M. P. Ferla, M. Garcia-Borràs, S. Osuna, and M. T. Reetz
“Pervasive cooperative mutational effects on multiple catalytic enzyme traits emerge via long-range conformational dynamics”
Nature Comm. 2021, 12, 1621 [abstract]
DOI: 10.1038/s41467-021-21833-w