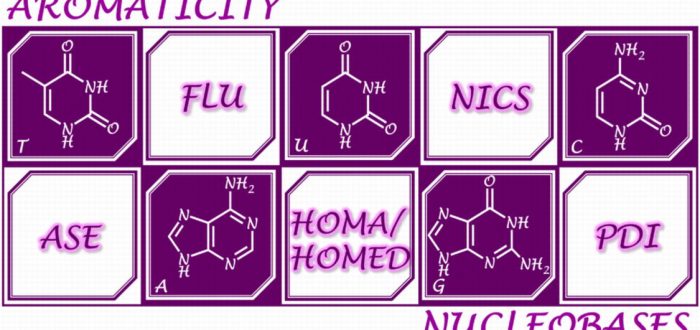
The 3D shape and the resulting physicochemical properties of double-helical DNA/RNA structures are determined not only by individual nucleobases, but also by their additive intermolecular interactions. Energetic contribution from aromatic pi-pi stacking to the stabilization of DNA/RNA is not small and sometimes even comparable to that from H-bonding. The basis of the stacking interactions lies in the pi-electron structure of individual nucleobases, which can be described by various aromaticity indices. Heteroatoms and exocyclic functional groups make the electronic structure of nucleobases different from aromatic hydrocarbons. Consequently, the cyclic pi-electron delocalization is not the only factor responsible for the relative stability of their tautomers. This review puts the spotlight on interplay between aromaticity of purine and pyrimidine nucleobases and their tautomeric preferences, as well as on the effects of different noncovalent interactions (hydrogen bonding, metal ion coordination, and pi–pi stacking) on pi-electron delocalization of five- and six-membered rings in individual nucleobases and their complexes.
The review was recently published in Wiley Interdisciplinary Reviews in Computational Molecular Science:
H. Szatylowicz, O.A. Stasyuk, M. Solà, and T.M. Krygowski
“Aromaticity of nucleic acid bases”
WIREs Comput Mol Sci 2020, [], ASAP-
DOI: 10.1002/wcms.1509