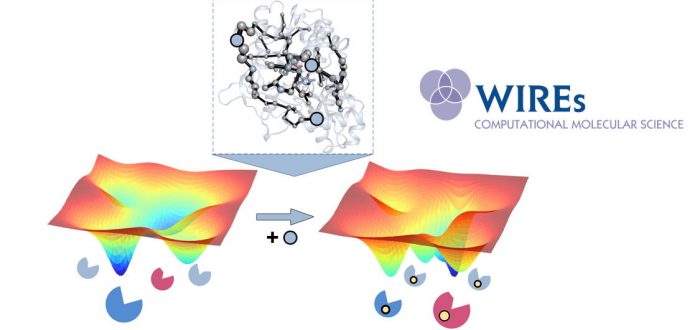
Many computational enzyme design approaches have been developed in recent years that focus on a reduced set of key enzymatic features. Initial protocols mostly focused on the chemical steps(s) through transition state stabilization, whereas most recent approaches exploit the enzyme conformational dynamics often crucial for substrate binding, product release and allosteric regulation. The detailed evaluation of the conformational landscape of many laboratory-evolved enzymes has revealed dramatic changes on the relative stabilities of the conformational states after mutation, favouring those conformational states key for the novel functionality. Of note is that these mutations are often located all around the enzyme structure, which contrasts with most of the computational design strategies that reduce the problem into active site alterations.
As shown in recent papers by the Osuna lab, computational strategies that consider enzyme design as a population shift problem, i.e. redistribution of the relative stabilities of the conformational states induced by mutations can be developed. These strategies focus on reconstructing the conformational landscape of the enzyme, and applying correlation-based tools (such as our in-house Shortest Path Map tool) to elucidate the underlying allosteric network of interactions and identify potential mutation hotspots located at the active site, but most importantly at distal positions for the first time.
This advance review by Osuna published in WIRES Computational Molecular Science provides a brief overview of the available computational enzyme design strategies, and focuses on highlighting some recent computational enzyme design protocols not restricted to active site mutations whose main focus is the alteration of enzyme conformational dynamics. The paper received really good referee reports which coincide that this is an excellent contribution that is likely to become an important reference for the field.
The review was recently published in Wiley Interdisciplinary Reviews in Computational Molecular Science:
S. Osuna
“The challenge of predicting distal active site mutations in computational enzyme design”
WIREs Comput Mol Sci 2020, ASAP
DOI: 10.1002/wcms.1502
Girona, September 10, 2020
More information: gestor.iqcc@gmail.com