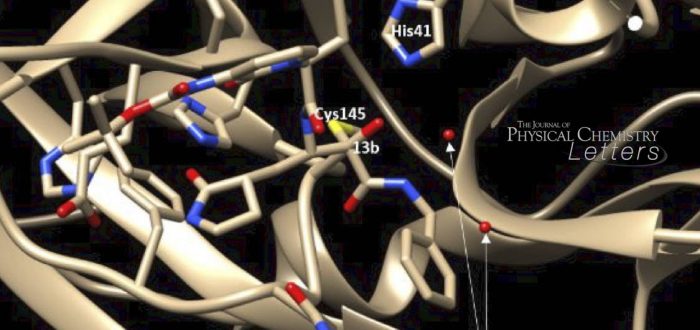
Recently, Hilgenfeld and co-workers reported (Science 2020, 368, 409–412) that alpha-ketoamide-derived substrates act as inhibitors of COVID by targeting the SARS-CoV-2 main protease. Their crystal structure analysis provided the basis for an in-depth computational study by Albert Poater, Serra Hunter fellow at IQCC (Univ. Girona). His calculations based on Density Functional Theory provided the origin for how the inhibitor produces a thermodynamically favorable interaction, and how it could be achieved kinetically. It was observed that a histidine, found in close proximity to the place where carbon-sulfur bond formation takes place, is key to understanding the mechanism of action of the inhibitor, which involves a proton transfer in the process. These findings encourage the search for new inhibitors (medicinal drugs) to fight the action of the virus, preferably without side-effects for humans. The research study is part of a larger project that received funding at the Red Española de Supercomputación in the form of computing hours at the BSC Mare Nostrum supercomputer in Barcelona, involving researchers from the IQCC (Javier Iglésias, Miquel Solà, Sílvia Osuna, Lluís Blancafort, Sílvia Simon, Albert Poater), UB (Jordi Poater) and Gran Canaria (Luis Miguel Azofra).
The paper was published today in Journal of Physical Chemistry Letters:
A. Poater
“Michael Acceptors Tuned by the Pivotal Aromaticity of Histidine to Block COVID19 Activity”
J. Phys. Chem. Lett. 2020, [], ASAP-
DOI: 10.1021/acs.jpclett.0c01828