Sílvia Osuna received her PhD in 2010 from the University of Girona (UdG) at the Institut de Química Computacional (IQC). In 2010, she moved to the group of Prof. Houk at the University of California, Los Angeles (UCLA) with a Marie Curie postdoctoral fellowship. She obtained a JdC postdoctoral contract at the UdG, a 5-year RyC contract (RYC-2014-16846), and in 2018 an ICREA Research position.
The group of Dr. Osuna was established thanks to the awarded 2015 European Research Council project – Starting grant project (ERC-2015-StG-679001) and focuses on the development of new computational tools and approaches for computational enzyme design. Her group is currently funded by a European Research Council project – Consolidator Grant, two ERC- Proof of Concept grants, and two I+D Spanish MINECO projects.
She been recently awarded the 2023 Young Spanish National Award in Chemistry (María Teresa Toral), 2021 EuChemS lecture award, the Catalan National Research Award – Young Talent 2019 from Fundació Catalana de Recerca i Innovació (FCRi), the Young Researcher award by the Royal Spanish Society of Chemistry (RSEQ 2016), Research award by the Fundación Princesa de Girona (FPdGi 2016- Science category), among many others.
- sec.iqcc@udg.edu
- +34 972 41 83 57
Osuna, Sílvia
Computational design of proficient enzymes: exploring the molecular basis of biocatalysis
Contact info:
Dr. Sílvia Osuna
silvia.osuna@udg.edu
Tel. (+34) 972 41 93 21
Website

Selected publications
Miguel A. Maria-Solano, Javier Iglesias-Fernández, Sílvia Osuna
Deciphering the Allosterically Driven Conformational Ensemble in Tryptophan Synthase Evolution
J. Am. Chem. Soc., 2019, 141, 13049-13056
DOI: 10.1021/jacs.9b03646
Christian Curado-Carballada, Ferran Feixas, Javier Iglesias-Fernández, Sílvia Osuna
Hidden Conformations inAspergillus niger Monoamine Oxidase are Key for Catalytic Efficiency
Angew. Chem. Int. Ed., 2019, 58, 3097-3101
DOI: 10.1002/anie.201812532
Xi Chen, Hongliu Zhang, Miguel A. Maria-Solano, Weidong Liu, Juan Li, Jinhui Feng, Xiangtao Liu, Sílvia Osuna, Rey-Ting Guo, Qiaqing Wu, Dunming Zhu, Yanhe Ma
Efficient reductive desymmetrization of bulky 1,3-cyclodiketones enabled by structure-guided directed evolution of a carbonyl reductase
Nat Catal, 2019, 2, 931-941
DOI: 10.1038/s41929-019-0347-y
Xavier Arqué, Adrian Romero-Rivera, Ferran Feixas, Tania Patiño, Sílvia Osuna, Samuel Sánchez
Intrinsic enzymatic properties modulate the self-propulsion of micromotors
Nat Commun, 2019, 10, 2826
DOI: 10.1038/s41467-019-10726-8
Antonia Tomás-Loba, Elisa Manieri, Bárbara González-Terán, Alfonso Mora, Luis Leiva-Vega, Ayelén M. Santamans, Rafael Romero-Becerra, Elena Rodríguez, Aránzazu Pintor-Chocano, Ferran Feixas, Juan Antonio López, Beatriz Caballero, Marianna Trakala, Óscar Blanco, Jorge L. Torres, Lourdes Hernández-Cosido, Valle Montalvo-Romeral, Nuria Matesanz, Marta Roche-Molina, Juan Antonio Bernal, Hannah Mischo, Marta León, Ainoa Caballero, Diego Miranda-Saavedra, Jesús Ruiz-Cabello, Yulia A. Nevzorova, Francisco Javier Cubero, Jerónimo Bravo, Jesús Vázquez, Marcos Malumbres, Miguel Marcos, Sílvia Osuna & Guadalupe Sabio
p38Υ is essential for cell cycle progression and liver tumorigenesis
Nature, 2019, 568, 557-560
DOI: 10.1038/s41586-019-1112-8
Dr. Sílvia Osuna
Research overview
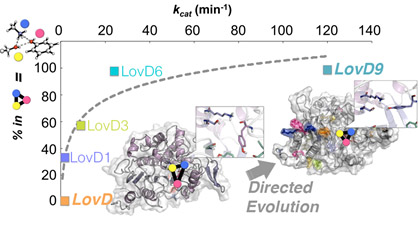
Billions of years of evolution have made enzymes superb catalysts capable of accelerating reactions by as many as seventeen orders of magnitude. This rate acceleration is achieved by decreasing the activation barriers of reactions, making them possible at lower temperatures and pressures. Enzymes (i.e. biocatalysts) are indeed the most efficient, specific and selective catalysts known. They operate under biological conditions, are biodegradable, non-toxic, their high selectivities and efficiencies reduce the number of work- up steps, and provide product in higher yields. These characteristics make enzyme-catalyzed processes an attractive alternative for chemical manufacturing. However, the use of enzymes in industry is limited, as most of processes do not present a biocatalyst to catalyze and accelerate the corresponding reactions. The ability of routinely designing enzymes for any target process will have large socio-economic impacts, as the production costs of many drugs will be reduced and will allow industries to use environmentally friendly alternatives. However, the routine design of enzymes for any target reaction has not yet been achieved. This is in part motivated by the imprecise knowledge of the underlying physical principles of biocatalysis, which makes the alteration of the natural activity of enzymes towards synthetically relevant targets a tremendous challenge for biochemistry. Current computational and experimental approaches are able to confer natural enzymes new functionalities but are economically unviable and the catalytic efficiencies lag far behind their natural counterparts.
We work in the design of new enzymes for distinct processes important for their potential applications in medicine. We explore the structural basis of improved catalysis achieved by the experimental directed evolution (DE) technique through computational modeling, and are currently developing a new computational protocol based on Molecular Dynamics and network models that reduce the complexity of the enzyme design paradigm. Our computational predictions are tested in the lab to finally elucidate the potential of this genuinely new computational approach for mimicking Nature’s rules of evolution.
We collaborate with many groups, being the most relevant ones: Prof. K. N. Houk (UCLA, USA), Prof. Y. Tang (UCLA, USA), Dr. G. Huisman (Codexis).
We additionally work on the computational exploration of the chemical reactivity and properties of carbon-based materials. This topic is related to Dr. Osuna’s PhD thesis and she has collaborations with the groups of Prof. L. Echegoyen (UTEP), Dr. Y. Yamakoshi (ETH Zurich), Prof. J. M. Poblet (URV), and Prof. N. Martín (UCM).
People
Principal Investigator
Staff and Postdocs

Alexander Swoboda
Postdoc
Supervisor:- S. Osuna

Eduard Masferrer
Postdoc
Supervisor:- S. Osuna

Guillem Casadevall
Postdoc
Supervisor:- S. Osuna
PhD and MACMoM students

Akram Doustmohammadi
PhD student
Supervisor:- S. Osuna

Cristina Duran
PhD Student (FPI)
Supervisor:- S. Osuna

Esther Pruna
PhD student
Supervisor:- S. Osuna

Janet Sánchez
PhD (FPI) student
Supervisor:- S. Osuna

Javier Moreno
PhD student
Supervisor:- S. Osuna

Jonnely Rissell Luizaga
PhD student
Supervisor:- S. Osuna
- M. Swart

Laia Ramoneda Giralt
PhD student
Supervisor:- S. Osuna

Rupal Sharma
PhD Student (IF-UdG)
Supervisor:- M. Swart
Funding
ERC
Researcher: Dr. Sílvia Osuna
Reference: ERC-2022-CoG-101088032
Funding: 1.996.250 €
Period: 01/10/2023 – 30/09/2028
ERC Proof of concept
Researcher: Dr Sílvia Osuna
Reference: ERC-2023-POC-101158166
Funding: 150.000 €
Period: 01/07/2024– 31/12/2025
Juan de la Cierva
Researcher: Dr. Eduard Masferrer (S. Osuna))
Reference: JDC2023-052689-I
Funding: – €
Period: 01/03/2025-28/02/2027
MCIU Proyectos I+D
Researcher: Dr. Sílvia Osuna
Reference: PDC2022-133950-100
Funding: 143.750 €
Period: 01/12/2022 – 31/12/2024
Researcher: Dr. Sílvia Osuna
Reference: PID2021-129034NB-100
Funding: 157.300 €
Period: 01/09/2022 – 31/08/2025
Marie Skłodowska-Curie
Researcher: Dr. Sílvia Osuna
Reference: HORIZON-MSCA-2023-DN-101169327
Funding: 2.563.653,60 € (UdG: 251.971,20€)
Period: 01/03/2025 – 28/02/2029
Researcher: Prof. Sílvia Osuna
Reference: HORIZON-MSCA-2024-DN- 101226960
Funding: 4.470.727,68 € (UdG: 282.188,16€)
Period: 2025-2029
Researcher: Prof. Sílvia Osuna
Reference: HORIZON-MSCA-2024-DN-JD-101226357
Funding: 3.869.142,72 € (UdG: 376.250,88 €)
Period: 2025-2030
AGAUR. Suport a grups de recerca.
Researcher: Dr. Sílvia Osuna
Reference: 2021 SGR 00487
Funding: 60.000 €
Period: 01/01/2022 – 31/12/2024
Collaborations
Nobu Tokuriki – University of British Columbia (Canada), collaboration with Sílvia Osuna.
Manfred Reetz – Max-Planck-Institut für Kohlenforschung (Germany), collaboration with Sílvia Osuna.
News
Sílvia Osuna receives IX Banc Sabadell Fundation award on Sciences and Engineering
Today, the Banc Sabadell Fundation announced that Prof. Sílvia Osuna, ICREA researcher at the Institut de
PhD defense Cristina Duran i Rebenaque
On 12th of June will be the defense of the doctoral thesis of Cristina
The need to implement FAIR principles in biomolecular simulations
In the Big Data era, a change of paradigm in the use of
MSCA Doctoral Network (COMENZE)
The project COMENZE: Computational and Experimental Enzyme Engineering for New Polymers, within the