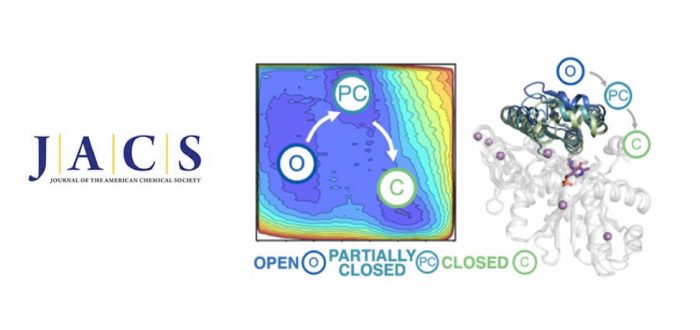
Multimeric enzyme complexes are ubiquitous in nature and catalyze a broad range of useful biological transformations. They are often characterized by a tight allosteric coupling between subunits, making them highly inefficient when isolated. A good example is Tryptophan synthase (TrpS), an allosteric heterodimeric enzyme in the form of an abba complex that catalyzes the biosynthesis of L-tryptophan. In this study, we decipher the allosteric regulation existing in TrpS from Pyrococcus furiosus (PfTrpS), and how the allosteric conformational ensemble is recovered in laboratory-evolved stand-alone ?-subunit variants. We find that recovering the conformational ensemble of a subdomain of TrpS affecting the relative stabilities of open, partially closed, and closed conformations is a prerequisite for enhancing the catalytic efficiency of the b-subunit in the absence of its binding partner. The distal mutations resuscitate the allosterically driven conformational regulation and alter the populations and rates of exchange between these multiple conformational states, which are essential for the multistep reaction pathway of the enzyme. Interestingly, these distal mutations can be a priori predicted by careful analysis of the conformational ensemble of the TrpS enzyme through computational methods. Our study provides the enzyme design field with a rational approach for evolving allosteric enzymes toward improved stand-alone function for biosynthetic applications. It was published recently in Journal of the American Chemical Society.
M.A. Maria-Solano, J. Iglesias-Fernández, and S. Osuna
“Deciphering the Allosterically Driven Conformational Ensemble in Tryptophan Synthase Evolution”
J. Am. Chem. Soc. 2019, 141, 13049-13056 [abstract]
DOI: 10.1021/jacs.9b03646